
Currently, the limitations in spatial resolution for in vivo finger MR imaging are subject motion, RF coil sensitivity and gradient strength. Further improvement of the spatial resolution such as in might facilitate the segmentation of finer structures. The spatial resolution of the MR images underlying this study is 195 × 195 × 200 µm 3, and the segmentation of a large variety of structures was possible, from the fingertip to approximately the center of the proximal phalanx.
Transmittance and reflectance images were formed by adding the weights of packets exiting the model in the forward or backward direction, respectively. A mirror boundary condition was applied whenever the model geometry extended beyond the limits of the simulated volume. Photon packets were stopped when their weight fell below a certain threshold. According to the simulated optical properties (absorption and scattering coefficients, refractive index and anisotropy), the photons were reflected, refracted or transported at voxel boundaries, and a fraction of the packet’s weight was absorbed in each voxel. Packets of photons with a weight of one were generated in the form of a perfectly collimated light source uniformly distributed over the whole geometry and transported through the simulated model. Briefly, the algorithm assumed homogeneity of optical properties of the tissue within each voxel of the model. Details of the custom simulation are presented in our previous work.
#Paraview image stack software#
The custom-made software was based on the Monte Carlo model of steady-state light transport in multilayered tissue (MCML), a well-established and validated code in the field of biomedical optics, which we extended for computations in fully 3D voxelated geometry and for parallel execution via several processor cores of a graphics card. To show the utility of the model, we applied a custom-weighted photon 3D Monte Carlo simulation of optical transport through the human index finger. As the model includes a number of tissue types, it may present a solid foundation for future simulations of various musculoskeletal disease processes in human joints. An example illustrating the utility of the model in biomedical applications is shown. High-resolution magnetic resonance images along with careful segmentation proved useful in the construction of an anatomically accurate model of the human index finger. The model was applied to the conditions of arthritic joint, ruptured tendon and variations in the geometry of a finger. In total, 15 TTs were identified: subcutis, Pacinian corpuscle, nerve, vein, artery, tendon, collateral ligament, volar plate, pulley A4, bone, cartilage, synovial cavity, joint capsule, epidermis and dermis. The final segmented model of the human index finger had a spatial resolution of 0.2 mm and included 6,809,600 voxels.
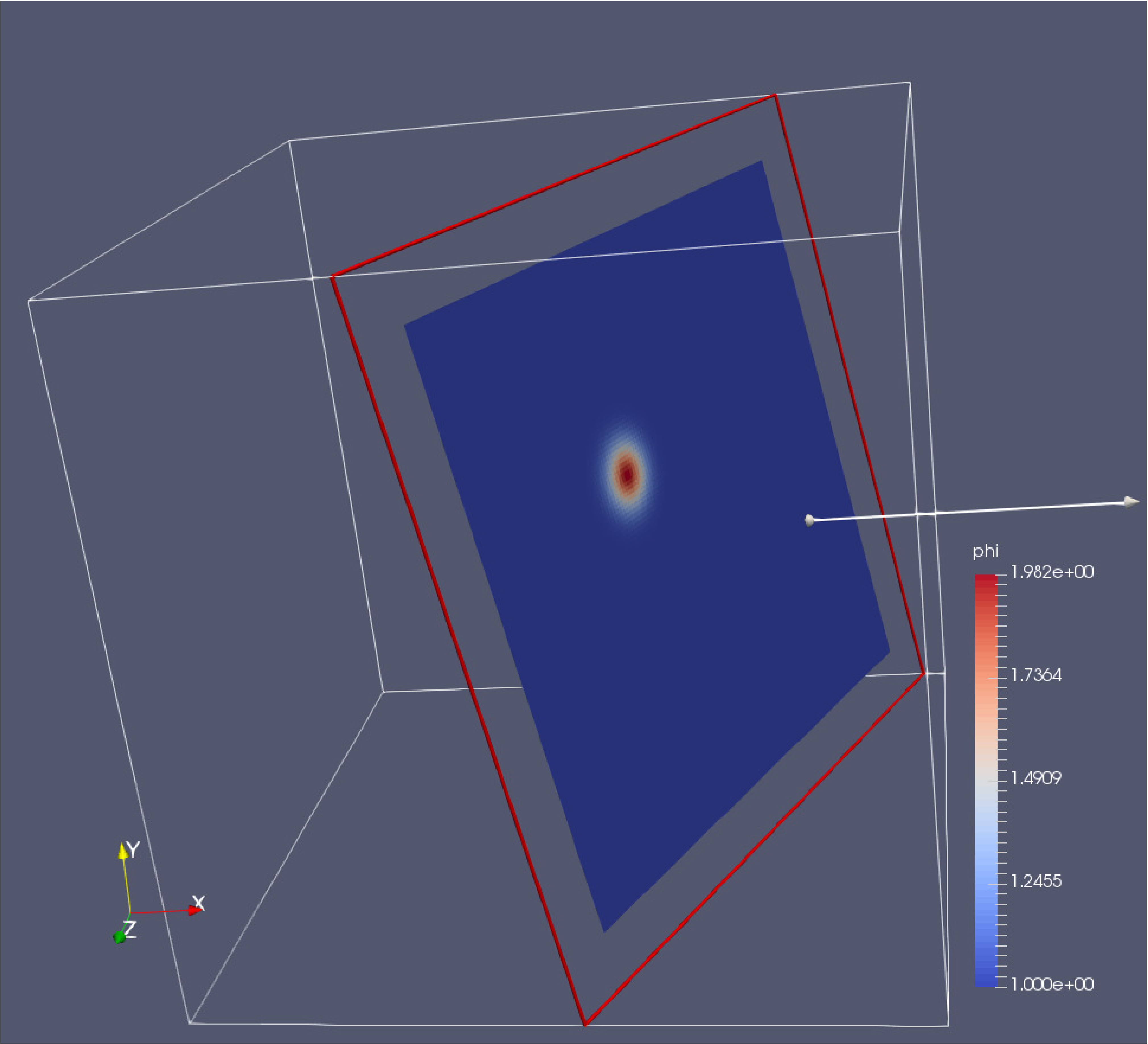
Segmentation was carried out under the supervision of an experienced radiologist to accurately capture various tissue types (TTs). Magnetic resonance images were acquired at ultrahigh magnetic field (7 T) with a radio-frequency coil specially designed for finger imaging.
In order to have potential clinical value, such models need to include a large number of tissue types, identified by an experienced professional, and should be versatile enough to be readily tailored to specific pathologies.

Anatomically accurate models of a human finger can be useful in simulating various disorders.


 0 kommentar(er)
0 kommentar(er)
